| Location: Home > Research > Research Progress |
| Researchers Chart Advances in Ancient DNA Technology |
|
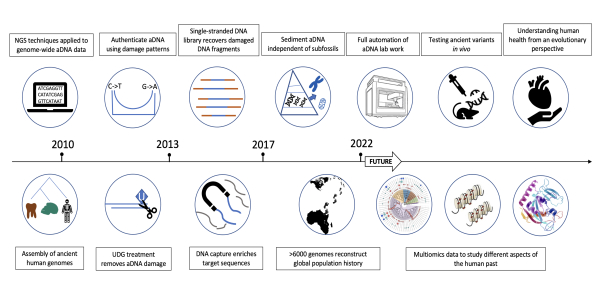
Over the past 10 years, researchers led by FU Qiaomei from the Institute of Vertebrate Paleontology and Paleoanthropology (IVPP) of the Chinese Academy of Sciences (CAS) have used ancient DNA (aDNA) technology to unearth the history of ancient human populations, especially those in East Asia. As part of their effort, the researchers reconstructed the whole genome of two extinct groups of archaic humans—Neanderthals and Denisovans; mapped the history of global population migrations and interactions; uncovered the genetic structure of the oldest East Asians; revealed adaptive genetic changes in East Asian Ice Age populations; and traced the formation of population patterns in northern and southern China as well as the origin of the Austronesian population in southern China. Recently, FU’s team reviewed the history of aDNA technological development, discussed current technical bottlenecks and solutions, and assessed the future of the technology. The study was published in Cell on July 21. A key technological development discussed in the study is high-throughput sequencing, which is a technique for rapidly sequencing large amounts of DNA. It can theoretically sequence all DNA molecules in a sample. Before high-throughput sequencing became commonplace, the aDNA field relied on polymerase chain reaction (PCR) techniques to sequence a few specific DNA fragments. Researchers could only extract a very limited amount of DNA information with this technology and had trouble distinguishing genuine aDNA from contaminant DNA. To complement advances in sequencing, aDNA researchers have also developed improved methods of DNA library construction to better reflect the characteristics of aDNA. Among these methods, partial uracil-DNA glycosylase (UDG) treatments and single-stranded DNA library construction are two of the most important. Partial UDG treatment not only preserves part of the DNA damage signal at DNA fragment tips but also removes most aDNA damage across the rest of the molecule. This method improves the accuracy of aDNA sequencing results while preserving aDNA features required for validation. Single-stranded DNA library construction allows the direct sequencing of damaged and denatured DNA fragments that may be lost in typical modern DNA library construction techniques. Advances in library construction have limited efficacy, however, since aDNA samples often contain a large amount of environmental DNA. As a result, useful endogenous aDNA sequences often account for less than 1% of the resulting sequences. To tackle this problem, researchers have applied DNA capture technology to the aDNA field by creating DNA and RNA probes with sequences similar to their targets. After adding the probes to sample extracts, the target aDNA binds to the probes and is then “fished out” from the massive amount of environmental DNA. This technology is widely used in ancient human genome research. Currently, more than two-thirds of ancient human genome data come from data captured using the “1240k” probe set. DNA capture technology not only greatly improves the efficiency of aDNA sequencing; it also allows recovery of usable data from samples that would otherwise be too degraded for analysis. More recently, aDNA researchers have pushed the envelope even further by extracting aDNA directly from “soil” (i.e., sediment). This technology has been applied to samples from the Denisova and Baishiya caves, making it possible to recover DNA from ancient humans who lived tens of thousands of years ago. Despite its fruitful results, however, the study of aDNA has always been very challenging. aDNA itself is highly susceptible to contamination, and experiments involving aDNA are extremely delicate. In the past, aDNA extraction and library construction were almost entirely dependent on manual operations. Recently, a few laboratories around the world have begun to integrate some aDNA methods with fully automated, pipetting robot platforms. However, at present, pre-processing of samples still requires manual steps. How to integrate this time-consuming and labor-intensive work into an automated system is the next challenge for aDNA experimental technology. The application of aDNA technology goes far beyond the ancient human genome, of course. Paleomolecular research also covers important topics such as tracing ancient epidemics and symbiotic microbial evolution through ancient microbial information; using ancient epigenetic information to explore the interaction between ancient animals and the environment; and using ancient proteins to explore human evolution over long time periods, including how aDNA influences the physiology and fitness of modern humans. aDNA is time-stamped genetic information that records the evolution and adaptation of human beings over tens of thousands of years. We now know from aDNA research that several important functional genetic haplotypes derive from archaic human populations. These genes are involved in innate immunity, lipid metabolism, high-altitude survivability, and skin color. However, the functions of most genetic variants identified by aDNA studies have not yet been determined. In the future, scientists may use the latest gene-editing technology to construct aDNA animal models that reveal the function of many unknown aDNA variants. This will help us better understand how modern-day human physiology and health have been affected by the genetic inheritance from our ancient forebears.
Link: https://doi.org/10.1016/j.cell.2022.06.009
Advances in aDNA technology since 2010 (Image by IVPP) |