An international team of researchers has reconstructed the common ancestor of placental mammals—an extremely diverse group including animals ranging from rodents to whales to humans—using the world’s largest dataset of both genetic and physical traits. In research published February 7 in the journal Science, the scientists reveal that, contradictory to a commonly held theory, placental mammals did not diversify into their present-day lineages until after the extinction event that eliminated non-avian dinosaurs, and about 70 percent of all species on Earth, some 65 million years ago.
This finding, and the visualization of the placental ancestor—a small, insect-eating animal— was made with the help of a powerful cloud-based and publicly accessible database called MorphoBank. The Science article is the result of a multi-year collaborative project funded by the National Science Foundation’s Assembling the Tree of Life program.
“Analysis of this massive dataset shows that placental mammals did not originate during the Mesozoic,” said lead author Maureen O’Leary, an associate professor in the Department of Anatomical Sciences in the School of Medicine at Stony Brook University and a research associate at the American Museum of Natural History. “Species like rodents and primates did not share the Earth with non-avian dinosaurs but arose from a common ancestor—a small, insect-eating, scampering animal—shortly after the dinosaurs’ demise.”
There are two major types of data for building evolutionary trees of life: phenomic data, observational traits such as anatomy and behavior; and genomic data encoded by DNA. Some scholars have argued that integration of both is necessary for robust tree-building because examining only one type of data (genomic or phenomic) leaves out significant information. The evolutionary history of placental mammals, for example, has been interpreted in very different ways depending on the data analyzed. One leading analysis based on genomic data alone predicted that a number of placental mammal lineages existed in the Late Cretaceous and survived the Cretaceous-Paleogene (KPg) extinction. Other analyses place the start of placental mammals near this boundary, and still others set their origin after this event.
“There are over 5,100 living placental species and they exhibit enormous diversity, varying greatly in size, locomotor ability, and brain size,” said Nancy Simmons, paper author and a curator in the Department of Mammalogy at the American Museum of Natural History. “Given this diversity, it’s of great interest to know when and how this clade first began evolving and diversifying.”
The new study combines genomic and phenomic data in a simultaneous analysis for a more complete picture of the tree of life.
“Despite the considerable contributions of DNA sequence data to the study of species relationships, phenomic data have a major role in the direct reconstruction of trees,” said author Michael Novacek, senior vice president, provost for science, and a curator of paleontology at the American Museum of Natural History. “Such data include features preserved in fossils where DNA recovery may be impossible. The mammalian record is notably enriched with well-preserved fossils, and we don’t want to build trees without using the direct evidence that these fossils contribute.”
“Discovering the tree of life is like piecing together a crime scene—it is a story that happened in the past that you can’t repeat,” O’Leary said. “Just like with a crime scene, the new tools of DNA add important information, but so do other physical clues like a body or, in the scientific realm, fossils, and anatomy. Combining all the evidence produces the most informed reconstruction of a past event.”
The tree of life produced in this study shows that placental mammals arose rapidly after the KPg extinction, with the original ancestor speciating 200,000-400,000 years after the event.
“This is about 36 million years later than the prediction based on purely genetic data,” said Marcelo Weksler, an author and research associate in the Museum’s Department of Mammalogy who is now at the Museu Nacional-UFRJ in Brazil.
The finding also contradicts a genomics-based model called the “Cretaceous-Terrestrial Revolution” that argues that the impetus for placental mammal speciation was the fragmentation of supercontinent Gondwana during the Jurassic and Cretaceous, millions of years earlier than the KPg event.
“The new tree indicates that the fragmentation of Gondwana came well before the origin of placental mammals and is an unrelated event,” said John Wible, paper author and curator of mammals at the Carnegie Museum of Natural History.
As part of the study, researchers used MorphoBank, an initiative funded primarily by the National Science Foundation (NSF) with additional support from Stony Brook University, the American Museum of Natural History, and the National Oceanic and Atmospheric Administration, to record phenomic traits for 86 placental mammal species, of which 40 were fossil species. The resulting dataset has more than 4,500 traits detailing characteristics such as the presence or absence of wings, teeth, and certain bones, type of hair cover, and structures found in the brain, as well as over 12,000 supporting images, all publicly available online. The dataset is an order of magnitude (10 times) larger than what has previously been used for studies of mammal relationships. Because phenomic datasets are built on physical objects like fossils that are limited in number and take time to excavate, prepare, and analyze, evolutionary trees based on anatomy usually don’t exceed several hundred traits. Large-scale collection of such data for tree-building is now being called “phylophenomics.”
“Cyberinfrastructure for organizing molecular biology has historically outstripped infrastructure for phenomic data, but new technologies like MorphoBank allow scientists working with phenomic data to produce larger and more complex projects and to enrich these databases with images, references, and comments,” said Andrea Cirranello, paper author and a postdoctoral research associate at Stony Brook University and the Museum.
The team reconstructed the anatomy of the placental common ancestor by mapping traits onto the tree most strongly supported by the combined phenomic and genomic data and comparing the features in placental mammals with those seen in their closest relatives. This method, known as optimization, allowed the researchers to determine what features first appeared in the common ancestor of placental mammals and also what traits were retained unchanged from more distant ancestors. The researchers conclude that the common ancestor had features such as a two-horned uterus, a brain with a convoluted cerebral cortex, and a placenta in which the maternal blood came in close contact with the membranes surrounding the fetus, as in humans.
In addition, the study reveals that a branch of the placental mammal tree called Afrotheria (because these animals—which range from elephants to aardvarks—live in Africa today) did not originate on that continent but rather in the Americas.
“Determining how these animals first made it to Africa is now an important research question along with many others that can be addressed using MorphoBank and the phylophenomic tree produced in this study,” said author Fernando Perini, a former postdoctoral fellow at the American Museum of Natural History who is now a professor at the Minas Gerais Federal University in Brazil.
Added author Mary Silcox, an assistant professor of anthropology at the University of Toronto Scarborough: “This project is not exhaustive, but exposes a way forward to collect data on other phenomic systems and other species.”
Other authors involved in this study include: Xijun Ni, Institute of Vertebrate Paleontology and Paleoanthropology (IVPP), Chinese Academy of Sciences; John Flynn, Andres Giallombardo, Norberto Giannini (and National University of Tucumánn, Argentina), Suzann Goldberg, Jin Meng (and IVPP), and Paul Velazco, American Museum of Natural History; Zhe-Xi Luo (now at University of Chicago) and Michelle Spaulding (and American Museum of Natural History), Carnegie Museum of Natural History; Jonathan Bloch and Zachary Randall, Florida Museum of Natural History; Guillermo Rougier, University of Louisville; Timothy Gaudin, University of Tennessee at Chattanooga; Brian Kraatz (and American Museum of Natural History), Western University of Health Sciences; Eric Sargis, Yale University, Peabody Museum of Natural History. (Reported by Kendra Snyder, American Museum of Natural History)

Fig.1 An artist’s rendering of the hypothetical placental ancestor, a small insect-eating animal. The research team reconstructed the anatomy of the animal by mapping traits onto the evolutionary tree most strongly supported by the combined phenomic and genomic data and comparing the features in placental mammals with those seen in their closest relatives. (Image by Carl Buell)
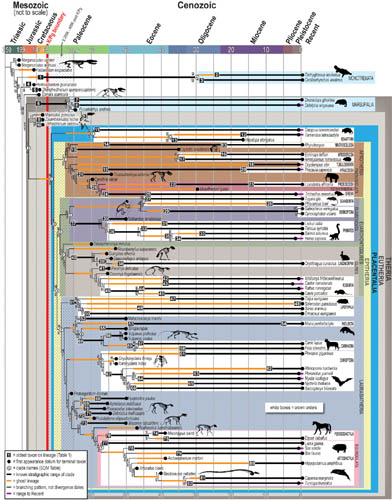
Fig.2 The new evolutionary tree for placental mammals, created by combining phenomic and genomic data. (Image by Stony Brook University/ Luci Betti Nash)
|

